Switching from Perl to Python: Speed
The job listings in scientific computing these days seem to show a mild preference for applicants with backgrounds in Python over Perl. It has high-profile (or just highly visible?) packages like NumPy and Python's MPI bindings for scientific computing, and some molecular dynamics packages (e.g., LAMMPS) include analysis routines written in Python. Although I've invested a few years into Perl, I've decided to not pigeonhole myself and start picking up Python. After all, Perl is unintelligible after it's been written, and it's sometimes frustrating to deal with its odd quirks.
To this end, I reimplemented one of my most-used Perl analysis routines in Python. Here is my Perl version, written back in 2009:
And here is the Python version I cooked up today:
In the Python version, there are several ways to tear through a file and I tried all three. Method #1 is closest to the Perl functionality, where I can specify multiple input files on the command line and have all of them parsed sequentially. Method #2 is the method that the Python documentation seems to advocate the most. Method #3 loads the whole file contents into memory and works from there.
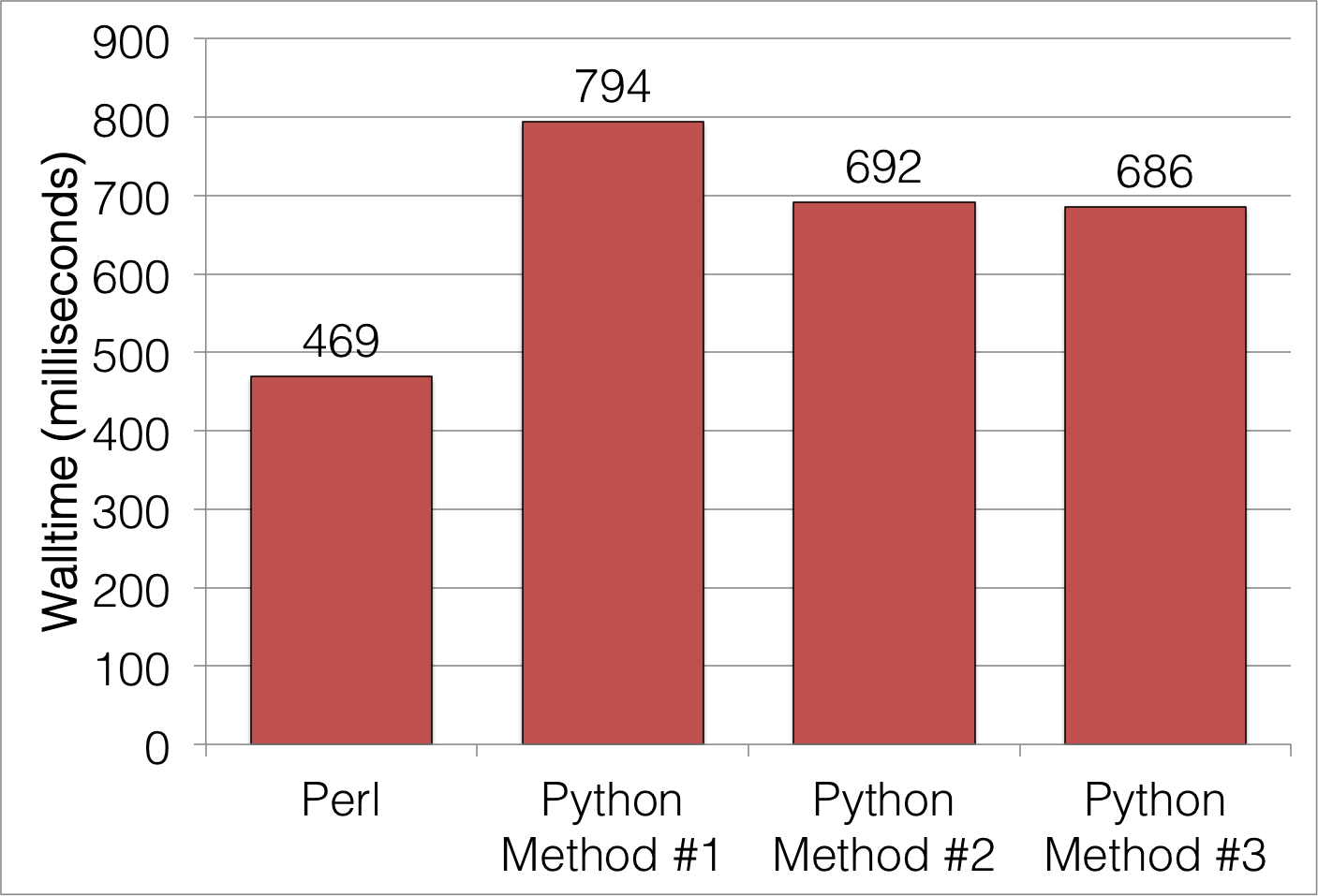
Unfortunately, in all three cases, Python seems to be slower than Perl. Average execution times for a typical input file are:
Maybe there's something I'm missing in the Python version, but the Perl version isn't exactly a shining example of simplicity in itself. What gives here? For a language that's being venerated in the scientific computing world, in the case of basic text parsing of large files, it isn't shining. At best, it's almost 50% slower than Perl.
To this end, I reimplemented one of my most-used Perl analysis routines in Python. Here is my Perl version, written back in 2009:
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| #!/usr/bin/perl | |
| @show = qw/ Siloxane SiO4 Si3O SiO3 SiO2 SiO1 NBO FreeOH H2O H3O SiOH SiOH2 Si2OH/; | |
| printf("\n%-8.8s ", "ird"); | |
| foreach $specie ( @show ) | |
| { | |
| printf("%8.8s ", $specie); | |
| } | |
| print "\n"; | |
| $current = 0; | |
| $isave = 0; | |
| while ( $line = <> ) | |
| { | |
| chomp($line); | |
| $line =~ s/^\s+//g; | |
| @arg = split(/\s+/, $line); | |
| next unless $line =~ m/^\d+\s+[\d\w]+\s+\d+\s+[\w\.]+\s+[\w\.]+\s+[\w\.]+\s*$/o; | |
| if ( $current == 0 ) | |
| { | |
| $current = $arg[0]; | |
| $isave = $current; | |
| } | |
| if ( $arg[0] != $current ) | |
| { | |
| &printargs(); | |
| $current = $arg[0]; | |
| $isave++; | |
| } | |
| $type{$arg[1]}++; | |
| } | |
| &printargs(); | |
| sub printargs( ) | |
| { | |
| printf("%-8s ", $isave); | |
| foreach $specie ( @show ) | |
| { | |
| printf("%8d ", $type{$specie}); | |
| } | |
| print "\n"; | |
| foreach $i ( keys(%type) ) | |
| { | |
| $type{$i} = 0; | |
| } | |
| } |
And here is the Python version I cooked up today:
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| #!/usr/bin/env python2 | |
| import fileinput | |
| import re | |
| show = [ "Siloxane", "SiO4", "Si3O", "SiO3", \ | |
| "SiO2", "SiO1", "NBO", "FreeOH", \ | |
| "H2O", "H3O", "SiOH", "SiOH2", "Si2OH" ] | |
| def printargs( counts, isave ): | |
| print "%-8s" % isave, | |
| for s in show: | |
| print "%8d" % counts[s], | |
| counts[s] = 0 | |
| print "\n", | |
| print "%-8s" % "ird", | |
| counts = {}; | |
| for s in show: | |
| counts[s] = 0 | |
| print "%8s" % s, | |
| print "\n", | |
| isave = 0; | |
| current = 0; | |
| RE_LINE = \ | |
| re.compile(r'\s*(\d+)\s+([\d\w]+)\s+\d+\s+[\w\.]+\s+[\w\.]+\s+[\w\.]+\s*$') | |
| for line in fileinput.input(): | |
| # for line in file('coord.out'): | |
| #contents = file('coord.out').readlines() | |
| #for line in contents: | |
| match = re.match(RE_LINE, line) | |
| if not match: continue | |
| specie = match.group(2) | |
| icur = int(match.group(1)) | |
| if current == 0: | |
| current = icur | |
| isave = current | |
| elif current != icur: | |
| printargs(counts, isave) | |
| current = icur | |
| isave += 1 | |
| if show.count(specie) > 0: | |
| counts[specie] += 1; | |
| printargs(counts,isave) |
In the Python version, there are several ways to tear through a file and I tried all three. Method #1 is closest to the Perl functionality, where I can specify multiple input files on the command line and have all of them parsed sequentially. Method #2 is the method that the Python documentation seems to advocate the most. Method #3 loads the whole file contents into memory and works from there.
Unfortunately, in all three cases, Python seems to be slower than Perl. Average execution times for a typical input file are:
Maybe there's something I'm missing in the Python version, but the Perl version isn't exactly a shining example of simplicity in itself. What gives here? For a language that's being venerated in the scientific computing world, in the case of basic text parsing of large files, it isn't shining. At best, it's almost 50% slower than Perl.